Traditional HPLC is Incapable of Reducing Cross Contamination of Custom Next-Generation Sequencing Adapters to Acceptable Levels
The development of next-generation massively parallel sequencing technologies (NGS), including the Roche 454™ Genome Sequencer FLX Instrument, the Illumina Genome Analyzer, Life Technology’s Ion Torrent™ Personal Genome Machine, and the Pacific Biosciences RS have revolutionized genomic and genetic research, significantly improved sequencing throughput, reduced costs for data production, and advanced research from weeks to hours.
Though there are different kinds of NGS technologies including sequencing by synthesis, sequencing by ligation, sequencing by hybridization, single molecule DNA sequencing, and nanopore sequencing, there are many similarities among the sequencing platforms. These can be grouped into library construction, library amplification, parallel sequencing, and data analysis. Synthetic oligonucleotides are used in each of these steps except in the data analysis stage. Each oligonucleotide type has a different set of manufacturing specifications needed to produce consistent results regardless of the sequencing instrument. Based on our experience and close collaboration with customers, we have found that the adapter sequences, which are ligated to sample DNA fragments and are used as both a way to bind fragments to the sequencing media and as an identification marker during multiplexing, carry the most stringent of specifications. The two most important aspects of adapter manufacturing are purity and levels of cross contamination.
In this note, we will focus on how traditional HPLC methods could lead to unwanted (yet undetected) levels of cross contamination, leading to false positives in multiplexing experiments–an expensive problem in terms of time and money.
Before one can determine if a manufacturing process meets minimum criteria for cross contamination, the proper analytical methods must be chosen. Kircher et. al. described a method for minimizing the degree of false calls coming from cross contamination during multiplexing, which is referred to as “double indexing”. (Martin Kircher, Susanna Sawyer, Matthias Meyer. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform, Nucleic Acids Research, 2012, e3-e3, DOI: 10.1093/nar/gkr771). In essence, the addition of indexes on both adapters used in an Illumina sequencer helped in determining which sequences belonged to which original sample of genomic DNA in spite of any pre-existing or caused contamination event where more than the desired index adapter was present. Using this same principle, we designed 12 unique synthetic oligonucleotides and manufactured them in isolation (to prevent contamination) in lieu of genomic DNA fragments to be sequenced. In this case, these sequences became the second index (each synthetic oligonucleotide had a predetermined base sequence) where the first index was one of Illumina’s pre-designed adapters. A total of 24 indexes were used for this experiment (Figure 1).
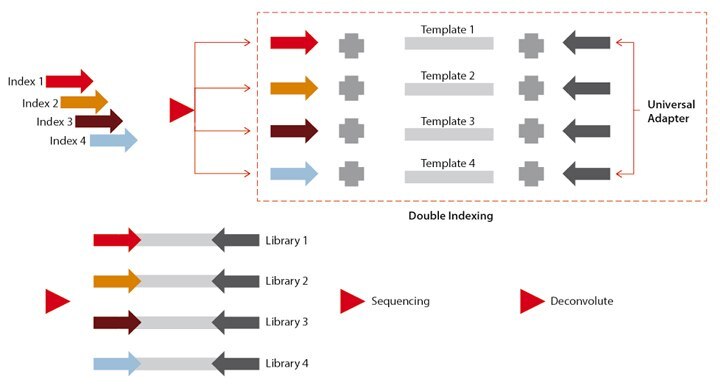
Figure 1. Experimental scheme using double indexing to identify potential cross contamination.
Each index was synthesized and purified using traditional HPLC conditions to recreate what a typical customer would receive from an oligonucleotide manufacturer and then use in a multiplexing experiment. Key to this set up was the fact that the adapter oligonucleotides were purified in sequential order and that the universal adapter was the last in the purification sequence (Table 1).

Table 1. Order of the HPLC purification of Illumina indexes used in this experiment.
After ligating purified adapters 1–12 to one of the unique 12 synthetic targets using currently accepted procedures for library construction, the double indexed construct was ready for sequencing. Once the sequencing data were collected and analyzed, the results were summarized on a matrix with the desired index on the horizontal axis and all the other potential contaminants on the vertical axis (Table 2).
Based on these experimental conditions, the analyzed data revealed that under traditional HPLC conditions, sequential purification of adapter oligonucleotides led to carryover contamination from the previously purified adapter, which ranged from 0.30–1.68% relative to the total number of sequencing reads (excluding the contribution of other contamination events). Furthermore, potential contamination from an adapter purified prior to the one immediately before the desired sequence could also be detected, though at much lower levels (0.02–0.17%). Additional care should be taken when also purifying the universal adapter as any contamination present on this oligonucleotide will carry over to all other indexes in the multiplex experiment. This experimental set up also revealed that there are other steps upstream and downstream of the purification process that could lead to carryover contamination. The latter is evident in the presence of contamination throughout all indexes of Index 6, 12, 14, 21, and 24 (Table 2).
The modification of the HPLC conditions as well as downstream and upstream steps to account for these observations helped produce adapter oligonucleotides which showed cross contamination levels below 0.3%. Since other factors can contribute to the observed cross-contamination percentage when the oligonucleotides are used for NGS sequencing that do not have to do with the manufacturing of the oligonucleotides (see reference before), it is paramount to have processes that minimize such events. Our engineers and scientists have used the above information to develop a set of manufacturing conditions to deliver oligonucleotides used in multiplexing experiments that can accommodate the different needs of customers using NGS.
To continue reading please sign in or create an account.
Don't Have An Account?