Binding and Kinetic Analysis of Protein Interactions Probed by Label-Free SpotMatrix SPR Technology in a Peptide Chip Format
Xue Y, Ferrell K, Monera O, Baggio R, Yan YX, Wassaf D, Barbisin M, Noble R, Palmer M, Arenas J, Hoge S, Gee MA
Protein-protein interactions play central roles in almost all cellular responses. Specific interactions between peptide motifs and signaling domains such as SH2 and WW domains are critical for a variety of signaling pathways. Many techniques have been developed to study protein binding to peptide motifs. These include ELISA, far western, immunoprecipitation or pull-down assays.
Applied Biosystems has developed a new platform that uses Grating-Coupled Surface Plasmon Resonance (GC-SPR) for kinetic measurement of molecular interactions between unlabeled analytes and biomolecules immobilized on affinity chip. This platform allows simultaneous affinity characterization of up to 400 targets spotted on a Gold Affinity Chip. We describe this SpotMatrix SPR technology and its application to protein-peptide interactions to illustrate how this technology is applied to the study of biomolecular interactions.
The SpotMatrix SPR Technology
The ABI SpotMatrix SPR platform uses a grating-coupled configuration, where a fine grating on the chip surface provides optical coupling and allows imaging of the entire surface at once, enabling simultaneous real-time binding analysis at every spot on the surface (Figure 1). The incident light hits the gold layer from the top and through the biomolecular layer, avoiding the stringent need for an optical quality support substrate and specific gold layer thickness.
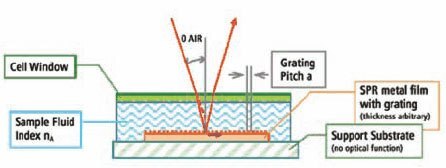
Figure 1.ABI SpotMatrix Instrument Schematic Schematic representation of the grating-coupled SPR configuration used in the ABI SpotMatrix instrumentation.
The SPR SpotMatrix Platform
Up to 400 targets can be spotted onto a 1 cm2 area of the affinity chip (Figure 2, left) using a standard floating-pin spotter. A 40 l flow cell is then assembled over the SpotMatrix by attachment of a selfadhesive window containing a fluid inlet and outlet (Figure 2, middle). The assembled affinity chip is then inserted into the instrument to measure the binding of an analyte to each of the immobilized targets using a CCD camera for imaging the entire surface in real time (Figure 2, right).

Figure 2.The SPR SpotMatrix Platform
Peptide Synthesis
Peptides were synthesized as 15 or 20 mers at Sigma using the PEPscreen® technology. The peptides contain an N-terminal biotin and a diethylene glycol linker between the biotin and the N-terminal amino acid residue. The average peptide purity for the 15 mer and 20 mer were 73% and 61%, respectively, and used without further purification.
Protein/Domain Binding to Peptide SpotMatrix
The biotinylated peptides were spotted onto a NeutrAvidin™ Affinity Chip using a Cartesian spotter. Analytes, including full-length proteins or domains, were flowed over the peptide SpotMatrix (Figure 3). Analyte binding to the peptides can be represented as end-point equilibrium binding (i.e., average of the last minute of binding signal before dissociation begins) as well as the affinity trace. Affinity constants, such as kon (ka), koff (kd), and kD (keq) were calculated using the Applied Biosystems 8500 Affinity Chip Analyzer data analysis software (Figure 4).
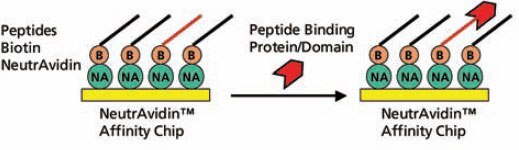
Figure 3.Schematic Diagram of the Assay
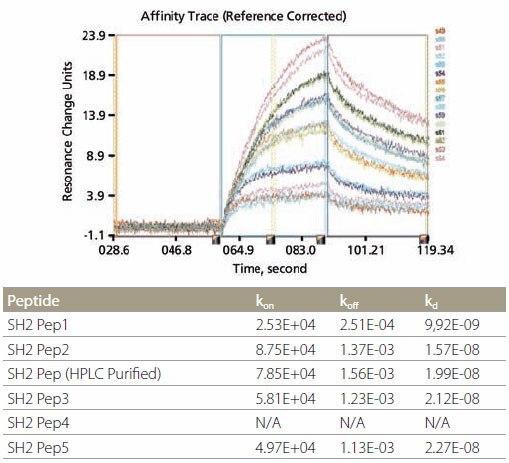
Figure 4.Kinetic Analysis and Data Example of kinetic analysis using SH2-binding peptides. Corresponding data are shown in the table above.
SH2 Domain-Peptide Interaction – Figure 5 demonstrates the specificity of binding between the peptides and two SH2 domains. The optimal binding motif for Grb2-SH2 domain is pYXNX, which is present in all 5 SH2 peptides containing phosphotyrosine (Figure 5, left). On the other hand, the optimal binding motif for SH2 domain of SH2-PTP2 is pY(N) X(V/I), which is present in SH2 Pep1 and SH2 Pep2 (Figure 5, right). All non-phosphorylated control peptides did not show any binding. Kinetic data for the binding of Grb2-SH2 binding are tabulated in Figure 4. It is noteworthy that the kinetic data for unpurified and HPLC-purified SH2 Pep2 are very similar.
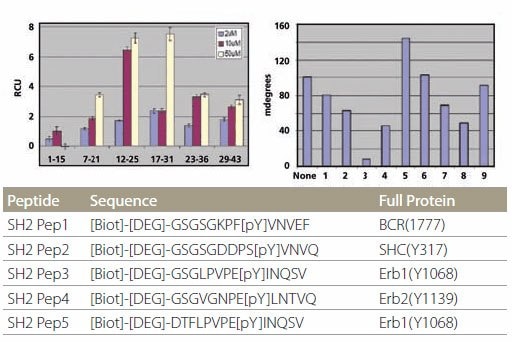
Figure 5.Specificity of SH2 Domain Building Specificity of SH2 domain binding. The peptides are plotted in the same order as the list on the table. Peptides designated with “C” are control peptides. The different color bars represent different protein concentrations.
Proline-rich Peptide Interaction with WW and SH3 Domains
Proline-rich peptide motifs are known to bind to a number of protein domains, such as SH3 and WW domains. WW domains can be further subdivided into four groups based on their particular binding preferences. Peptides representing all four binding motifs, along with a control peptide (Figure 6, table), were spotted onto the affinity chip. The WW domain of FBP21 protein specifically binds only to the SmB and P3 peptides that both belong to subclass III (Figure 6, left). On the other hand, the WW domain of GST-Nedd4 binds only to WBP1 peptide (Figure 6, right). These specificities agree with literature data.
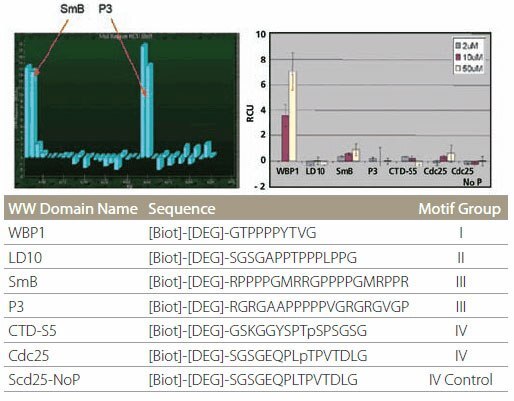
Figure 6.Specificity of WW Domain Binding Specificity of WW domain binding. The color bars represent different protein concentrations.
Mapping of the Actin Binding Site on Thymosin 4
Thymosin 4 is a ubiquitous 43 amino acid polypeptide that is an important mediator of cell proliferation, migration, and differentiation. It is believed to be one of the main G-actin sequestering peptides. It is also an anti-tumor target due to its important role in angiogenesis.
Six overlapping peptide sequences of thymosin 4 were synthesized using the Pepscreen platform and spotted into the SpotMatrix without further purification. Monomeric G-actin (8.7 M) was then flowed over the peptide affinity chip. Figure 7 (left) shows that peptides 12-26 and 17-31 have the highest binding activity to actin. Sequential alanine substitution of the amino acids in peptide 12-26 (Figure 7, right) showed that the lysine residue at position 18 is essential for actin binding.
Figure 7.End-point Binding Analysis Left: End-point binding of each overlapping peptide to G-actin. Right: Screening of essential amino acids in thymosin β412-26 peptide by Ala substitution in the positions indicated.
Modification-Specific Antibody Binding
Modification specific antibodies are widely used by researchers studying signal transduction pathways. However, their affinity and specificity should be carefully evaluated. The SpotMatrix SPR platform is well-suited for this application. For example, when different peptides were spotted on the SpotMatrix and a phosphotyrosine-specific antibody was flowed over the affinity chip, only the phosphotyrosine peptides showed binding to the antibody (Figure 8, left). A similar study using phosphothreonine-specific antibody also showed binding to both phosphotyrosine- and phosphothreonine-containing peptides (Figure 8, right). Non-phosphorylated peptides and phosphoserinecontaining peptides did not bind. The affinity of binding can also be determined, as previously mentioned.
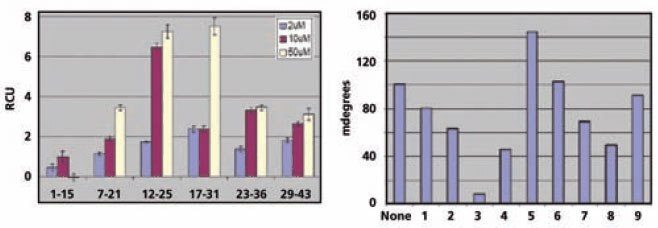
Figure 8.Binding Analysis of Antibodies to Immobilized Peptides Binding of phosphotyrosine-specific (left) and phosphothreonine-specific (right) antibodies to peptides immobilized on the affinity chip.
Conclusion
These studies demonstrated several examples of the combined power of combinatorial peptide libraries using the PEPscreen platform and SpotMatrix SPR analysis in interrogating specific protein-protein interactions. Among the benefits are in mapping the protein epitopes, determination of the binding constants, and simultaneous, real-time assays in a high throughput format.
To continue reading please sign in or create an account.
Don't Have An Account?