05-1242
Anti-trimethyl-Histone H3 (Lys9) Antibody, clone 6F12-H4
clone 6F12-H4, from mouse
Synonym(s):
H3K9me3, Histone H3 (tri methyl K9), H3 histone family, member T, histone 3, H3, histone cluster 3, H3
About This Item
Recommended Products
biological source
mouse
Quality Level
antibody form
purified antibody
antibody product type
primary antibodies
clone
6F12-H4, monoclonal
species reactivity
mouse, human
technique(s)
ChIP: suitable (ChIP-seq)
dot blot: suitable
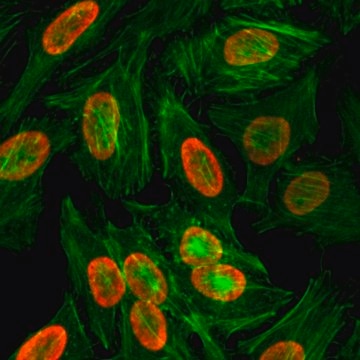
immunofluorescence: suitable
inhibition assay: suitable (peptide)
western blot: suitable
isotype
IgG1κ
NCBI accession no.
UniProt accession no.
shipped in
wet ice
target post-translational modification
trimethylation (Lys9)
Gene Information
human ... H3C1(8350)
mouse ... H3C1(360198)
Related Categories
General description
Specificity
Application
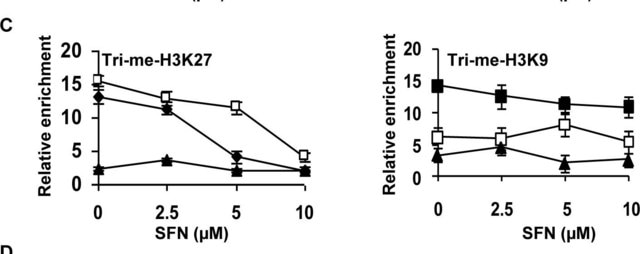
Representative data from a previous lot. Sonicated 3T3 L1 chromatin was subjected to chromatin immunoprecipitation using anti- trimethyl-histone H3 (Lys9) and the Magna ChIP G (Cat. #17-611) Kit. Successful immunoprecipitation of trimethylhistone H3 (Lys9) associated DNA fragments was verified by qPCR using primers flanking the p16 promoter.
Peptide Inhibition Analysis:
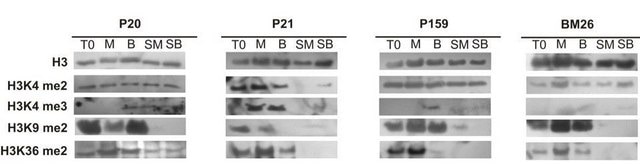
Peptide blocking assay demonstrates distinct preference of the antibody for the trimethyl form vs. the dimethyl form.
Chromatin Immunoprecipitation (ChIP):
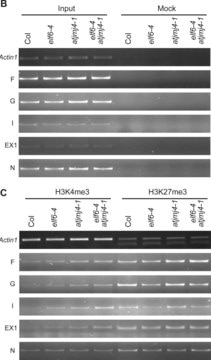
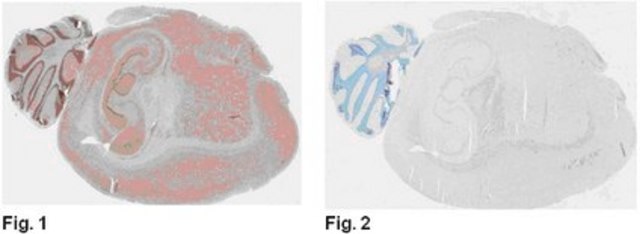
ChIP analysis of known chromosomal Suv39h targets (H3K9me3 in major satellites, mouseES cells).
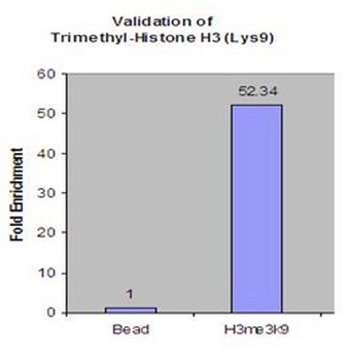
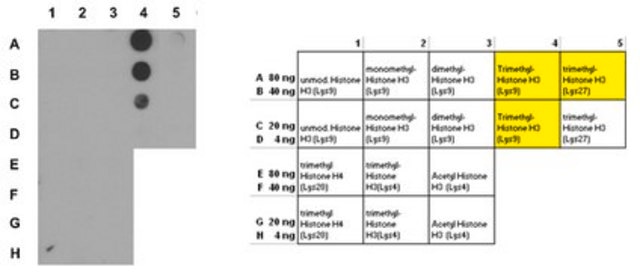
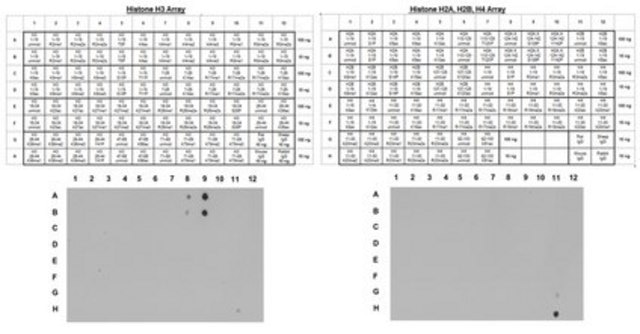
Dot Blot Analysis:
Dot-blot analysis demonstrating specificity of anti-H3K9me3, clone 6F12-H4 for trimethyl Lys9 of histone H3.
Quality
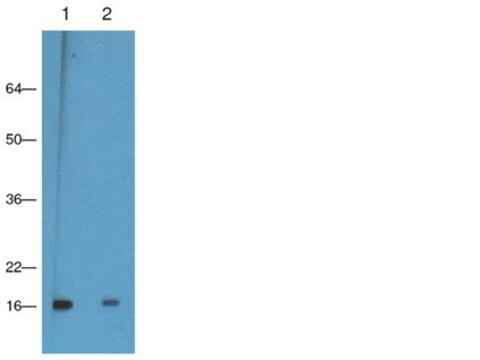
Western Blot Analysis: A 0.5 – 5 μg dilution of this lot detected trimethyl histone H3 (Lys9) in HeLa acid extracts.
Target description
Physical form
Not finding the right product?
Try our Product Selector Tool.
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service