412512
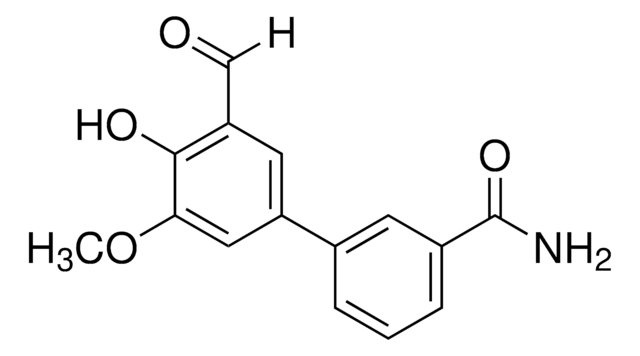
IRE1 Inhibitor III, 4μ8C
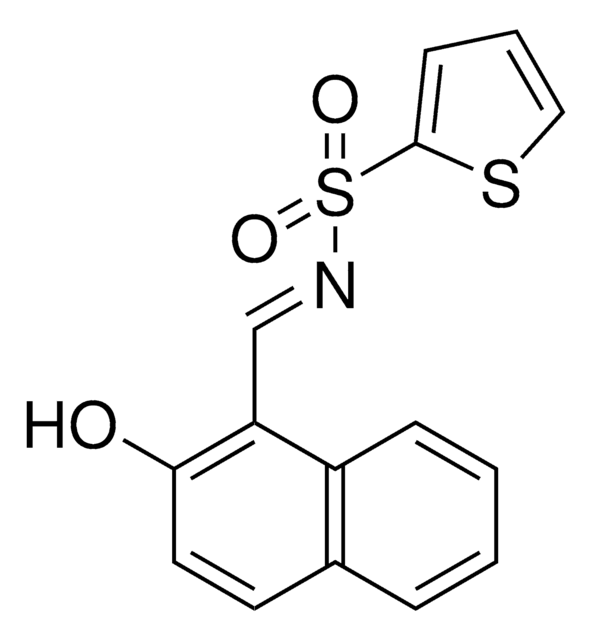
IRE1 Inhibitor III, CAS 14003-96-4, is a cell-permeable. Covalent inhibitor of IRE1 RNase activity (IC₅₀ = 550 and 45 nM, respectively, with 0 & 16 min preincubation in RNA cleavage assays).
Synonym(s):
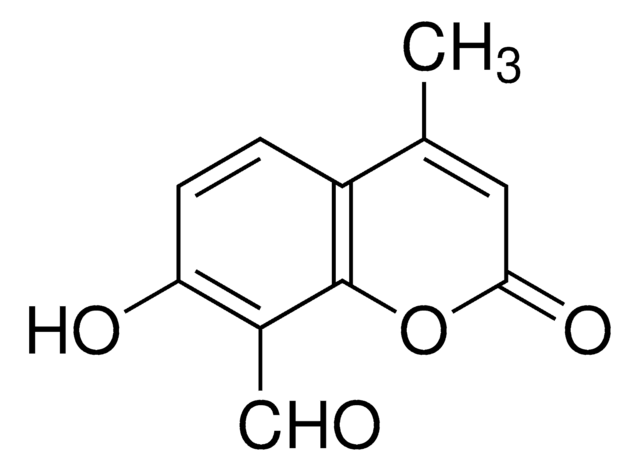
IRE1 Inhibitor III, 4μ8C, 8-Formyl-7-hydroxy-4-methylcoumarin, 7-Hydroxy-4-methyl-2-oxo-2H-chromene-8-carbaldehyde, ER-to-Nucleus Signaling 1 Inhibitor III, Inositol-Reguiring Protein 1 Inhibitor III
About This Item
Recommended Products
Quality Level
assay
≥95% (HPLC)
form
powder
manufacturer/tradename
Calbiochem®
storage condition
OK to freeze
protect from light
color
yellow
solubility
DMSO: 25 mg/mL
shipped in
ambient
storage temp.
2-8°C
SMILES string
O=C(C=C1C)OC2=C1C=CC(O)=C2C=O
InChI
1S/C11H8O4/c1-6-4-10(14)15-11-7(6)2-3-9(13)8(11)5-12/h2-5,13H,1H3
InChI key
RTHHSXOVIJWFQP-UHFFFAOYSA-N
General description
Biochem/physiol Actions
IRE1
Packaging
Warning
Reconstitution
Other Notes
Legal Information
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service
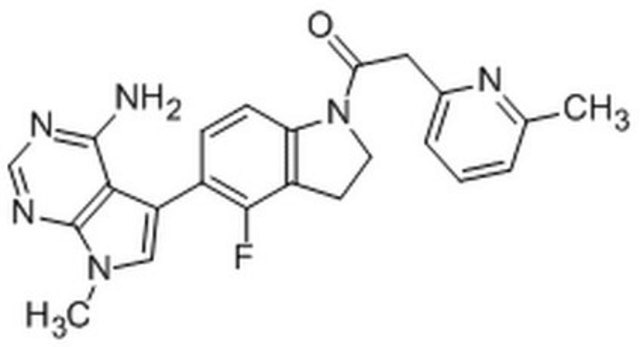
![PERK Inhibitor I, GSK2606414 GSK2606414 is a cell-permeable, highly potent inhibitor of EIF2AK3/PERK (IC₅₀ = 0.4 nM; [ATP] = 5 µM). Targets PERK in its inactive DFG conformation at the ATP-binding region.](/deepweb/assets/sigmaaldrich/product/structures/180/559/efa716dc-d5fe-4339-a6f0-0103084fc04a/640/efa716dc-d5fe-4339-a6f0-0103084fc04a.png)