11585606910
Roche
DIG-High Prime
sufficient for 40 labeling reactions, pkg of 160 μL, solution
Synonym(s):
DIG system
About This Item
Recommended Products
form
solution
Quality Level
usage
sufficient for 40 labeling reactions
packaging
pkg of 160 μL
manufacturer/tradename
Roche
greener alternative product characteristics
Designing Safer Chemicals
Learn more about the Principles of Green Chemistry.
greener alternative category
storage temp.
−20°C
General description
Specificity
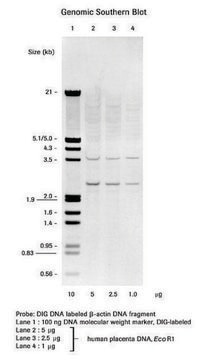
Application
- in Southern blots
- in northern blots
- in dot/slot blots
- for screening of gene libraries
- in In situ hybridizations
Features and Benefits
- DNA amounts ranging from 10ng to 3μg in a standard reaction.
- DNA of different lengths ranging from small restriction fragments to λ or cosmid DNA.
- DNA, supercoiled or linearized.
- DNA in low melting-point agarose.
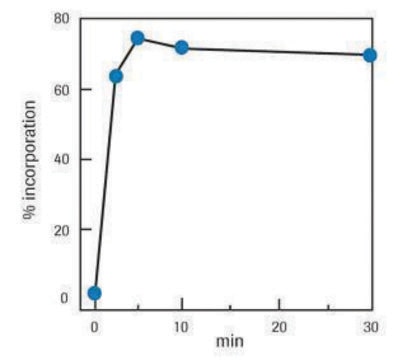
Labeling efficiency:
A standard labeling reaction with 1μg template yields 0.8μg newly synthesized digoxigenin-labeled DNA after 1 hour, and 2μg after a 20-hour incubation at +37°C.
Contents
5x concentrated random primer mix: 1U/ μl Klenow polymerase, labeling grade, 1mM dATP, 1mM dCTP, 1mM dGTP, 0.65mM dTTP, 0.35mM DIG-11-dUTP, alkali labile in 50% (v/v) glycerol.
Quality
In a standard assay with 1μg linearized pBR 328, 0.8μg of DIG-labeled DNA is synthesized after 1 hour, and 2.3μg after 20 hours. When this labeled DNA is used for hybridization at a concentration of 20ng/ml, 0.03pg homologous DNA are detected by chemiluminescence with the anti-DIG-alkaline phosphatase conjugate and CSPD on a dot or Southern blot.
Principle
Preparation Note
Assay Time: 80 minutes
Sample Materials
- DNA fragments of at least 100bp
- Linearized plasmid, cosmid or λDNA
- Supercoiled DNA
- Or minimal amounts of DNA (10ng), e.g., DNA restriction fragments isolated from gels or in molten agarose
Note: To obtain optimal results, template DNA should be linearized and should have a size of = 100bp or larger. Template DNA > 5kb should be restriction-digested using a 4bp cutter prior to labeling.
Other Notes
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
No data available
flash_point_c
No data available
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Articles
Digoxigenin (DIG) labeling methods and kits for DNA and RNA DIG probes, random primed DNA labeling, nick translation labeling, 5’ and 3’ oligonucleotide end-labeling.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service