11767291910
Roche
TUNEL Label Mix
sufficient for 30 tests, pkg of 3 × 550 μL
Synonym(s):
transferase dUTP nick end labeling, tunel
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
41105500
Recommended Products
form
solution
Quality Level
usage
sufficient for 30 tests
packaging
pkg of 3 × 550 μL
manufacturer/tradename
Roche
color
colorless
solubility
water: miscible
storage temp.
−20°C
Related Categories
General description
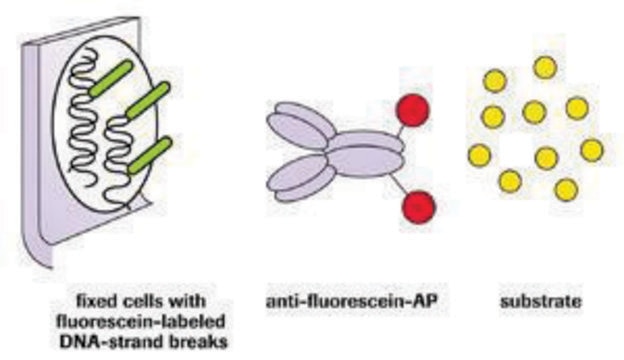
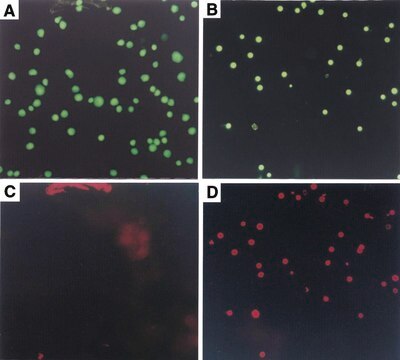
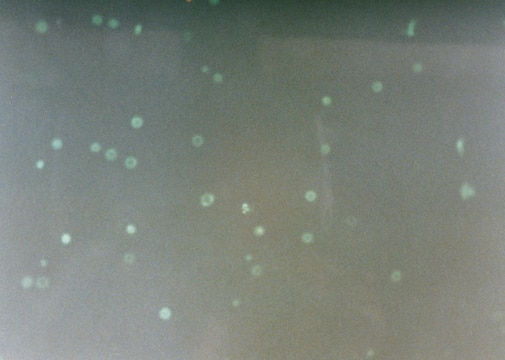
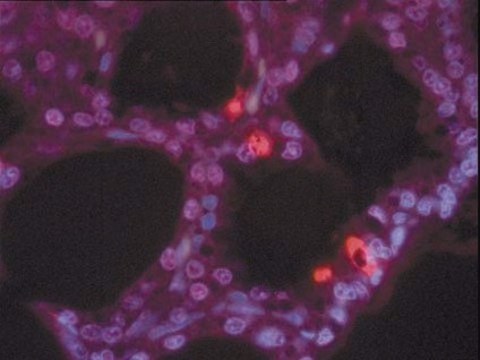
The nucleotide-labeling mix (TUNEL Label) contains fluorescein-dUTP and -dNTPs, both needed to perform the TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) reaction for the detection of apoptosis in situ. The nucleotide-labeling mix is used in combination with the TUNEL enzyme to prepare a TUNEL reaction mixture. This reaction mixture is used to label DNA-strand breaks for detecting and quantifying apoptotic cell death at a single-cell level in cells and tissues.
Application
TUNEL Label Mix has been used for the determination of apoptosis using TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay and DNA nick-end labeling by the TUNEL method.
The nucleotide-labeling mix (TUNEL Label) contains fluorescein-dUTP and -dNTPs, both needed to perform the TUNEL reaction for the detection of apoptosis in situ. The nucleotide-labeling mix is used in combination with the TUNEL Enzyme to prepare a TUNEL reaction mixture. This reaction mixture is used to label DNA-strand breaks for detecting and quantifying apoptotic cell death at a single-cell level in cells and tissues.
Preparation Note
Working solution: TUNEL Label is used in combination with the TUNEL Enzyme to prepare the TUNEL reaction mixture.
For one test: Mix 45 μl TUNEL Label with 5 μl TUNEL Enzyme prior to use. For negative control use 50 μl/test TUNEL Label only.
Storage conditions (working solution): Note: The TUNEL reaction mixture (45 μl TUNEL Label with 5 μl TUNEL Enzyme for 1 test) should be prepared just before use, and should not be stored. Keep the TUNEL reaction mixture on ice until use.
For one test: Mix 45 μl TUNEL Label with 5 μl TUNEL Enzyme prior to use. For negative control use 50 μl/test TUNEL Label only.
Storage conditions (working solution): Note: The TUNEL reaction mixture (45 μl TUNEL Label with 5 μl TUNEL Enzyme for 1 test) should be prepared just before use, and should not be stored. Keep the TUNEL reaction mixture on ice until use.
Other Notes
For life science research only. Not for use in diagnostic procedures.
signalword
Danger
hcodes
Hazard Classifications
Aquatic Chronic 2 - Carc. 1B Inhalation
wgk_germany
WGK 3
flash_point_f
does not flash
flash_point_c
does not flash
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Liang Liu et al.
Frontiers in oncology, 11, 648152-648152 (2021-08-13)
Glioma is the most common primary tumour of the central nervous system and is considered one of the greatest challenges for neurosurgery. Mounting evidence has shown that lncRNAs participate in various biological processes of tumours, including glioma. This study aimed
Ding Tian et al.
Cell death & disease, 11(7), 526-526 (2020-07-15)
Dysfunction of endothelial progenitor cells (EPCs) is a key factor in vascular complications of diabetes mellitus. Although the roles of microRNAs and circular RNAs in regulating cell functions have been thoroughly studied, their role in regulating autophagy and apoptosis of
Karl J Wahlin et al.
PloS one, 8(11), e79140-e79140 (2013-11-19)
Vertebrate genomes undergo epigenetic reprogramming during development and disease. Emerging evidence suggests that DNA methylation plays a key role in cell fate determination in the retina. Despite extensive studies of the programmed cell death that occurs during retinal development and
Yan Yang et al.
Neural regeneration research, 15(3), 464-472 (2019-10-02)
Mitochondrial dysfunction in neurons has been implicated in hypoxia-ischemia-induced brain injury. Although mesenchymal stem cell therapy has emerged as a novel treatment for this pathology, the mechanisms are not fully understood. To address this issue, we first co-cultured 1.5 ×
Sophie A Montandon et al.
EvoDevo, 5, 33-33 (2015-02-24)
Mammals exhibit a remarkable variety of phenotypes and comparative studies using novel model species are needed to uncover the evolutionary developmental mechanisms generating this diversity. Here, we undertake a developmental biology and numerical modeling approach to investigate the development of
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service