SRP0179
Lysyl Oxidase-like 2 (LOXL2) FLAG-tag (Sf9-derived) human
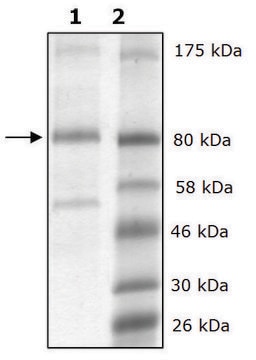
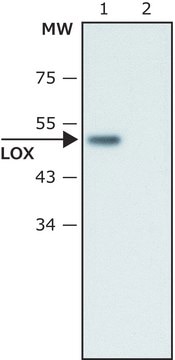
recombinant, expressed in baculovirus infected insect cells, ≥65% (SDS-PAGE)
Synonym(s):
LOR2, LOXL2, WS9-14
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
12352204
NACRES:
NA.32
Recommended Products
biological source
human
recombinant
expressed in baculovirus infected insect cells
assay
≥65% (SDS-PAGE)
form
aqueous solution
packaging
pkg of 10 μg
concentration
>0.02 mg/mL
NCBI accession no.
UniProt accession no.
shipped in
dry ice
storage temp.
−70°C
Gene Information
human ... LOXL2(4017)
General description
The gene LOXL2 (lysyl oxidase like 2) is mapped to human chromosome 8p21.3. It is widely expressed and belongs to the LOX family of proteins. The protein has a conserved C-terminus region with catalytic function and a divergent N-terminal part.
Human LOXL2 (GenBank Accession No. NM_002318), amino acids 500-end, MW = 32kDa, expressed in a Baculovirus infected Sf9 cell expression system.
Human LOXL2 (GenBank Accession No. NM_002318), amino acids 500-end, MW = 32kDa, expressed in a Baculovirus infected Sf9 cell expression system.
Application
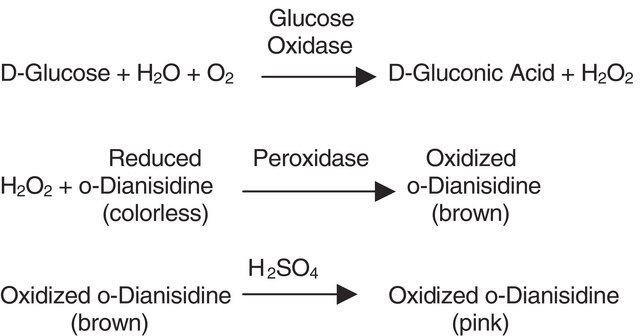
Useful for the study of enzyme kinetics, screening inhibitors, and selectivity profiling.
Biochem/physiol Actions
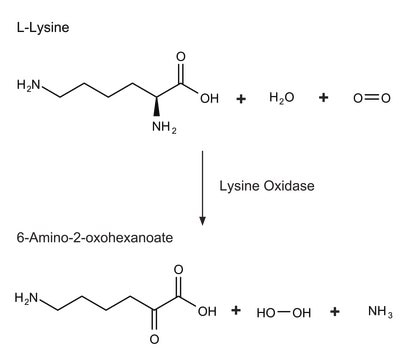
LOXL2 (lysyl oxidase like 2) is an amino oxidase which is dependent on copper and quinone. LOXL2 participates in various events, such as gene transcription, cell migration, adhesion, angiogenesis and differentiation. It is responsible for connecting collagen and/or elastin in the extracellular matrix. It deaminates trimethylated lysine 4 in histone H3, causing transcriptional repression. LOXL2 also oxidises methylated TAF10 (TATA-box binding protein associated factor 10), suppressing some TFIID (transcription factor IID)-associated genes. The LOXL2 gene is upregulated in various cancers, such as gastric cancer, breast cancer and squamous cell carcinomas. The gene is associated with cancer cell invasion, lymph node metastasis and poor overall patient survival.
Physical form
Formulated in 25 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.05% Tween-20, 20% glycerol and 3 mM DTT
Preparation Note
Thaw on ice. Upon first thaw, briefly spin tube containing enzyme to recover full content of the tube. Aliquot enzyme into single use aliquots. Store remaining undiluted enzyme in aliquots at -70°C. Note: Enzyme is very sensitive to freeze/thaw cycles.
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
The function and mechanisms of action of LOXL2 in cancer (Review).
Wu L and Zhu Y
International Journal of Molecular Medicine, 36, 1200-1204 (2015)
The LOXL2 gene encodes a new lysyl oxidase-like protein and is expressed at high levels in reproductive tissues.
Jourdan-Le Saux C, et al.
The Journal of Biological Chemistry, 274, 12939-12944 (1999)
LOXL2 Oxidizes Methylated TAF10 and Controls TFIID-Dependent Genes during Neural Progenitor Differentiation.
Iturbide A, et al.
Molecular Cell, 58, 755-766 (2015)
Rika Nishikawa et al.
FEBS letters, 589(16), 2136-2145 (2015-06-23)
Here, we found that members of the microRNA-29 family (miR-29a/b/c; "miR-29s") were significantly reduced in clear cell renal cell carcinoma (ccRCC) tissues, suggesting that they functioned as tumour suppressors. Restoration of all mature members of the miR-29 family inhibited cancer
Keiko Mizuno et al.
International journal of oncology, 48(2), 450-460 (2015-12-18)
Lung cancer remains the most frequent cause of cancer-related death in developed countries. A recent molecular-targeted strategy has contributed to improvement of the remarkable effect of adenocarcinoma of the lung. However, such treatment has not been developed for squamous cell
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service