CRISPR/CAS9 Gene Editing Protocol for Human Induced Pluripotent Stem Cells (iPSCs)
Introduction
Induced pluripotent stem cells (iPSCs), have the capacity to give rise to differentiated progeny arising from of all germ layers of the body including: ectoderm, endoderm, and mesoderm. The ability to expand patient derived human iPSCs in vitro and subject them to cell-type specific differentiation protocols is the basis for generating “disease-in-a-dish” cellular models for basic stem cell research and drug-discovery applications. Recently, gene editing technologies such as CRISPR/CAS9 have allowed the generation of isogenic control human iPS cell lines to study the genetic mechanism behind disease and cellular functionality. Relying on our extensive experience in both gene editing and stem cell culture, we have developed a step-by-step protocol guide that can be followed to perform gene editing in human iPSCs using CRISPR/CAS9 technology.
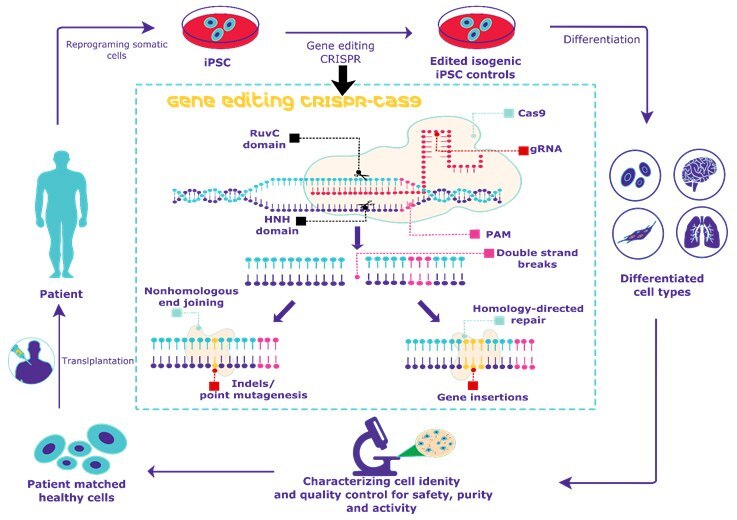
Figure 1. Overview of CRISPR gene editing of human iPSCs. Gene editing of human iPSCs using CRISPR/CAS9 allows for the generation of isogenic disease controls for stem cell research applications.
Methods
It is important to start with high quality iPS cells prior to initiating any CRISPR/Cas9 gene editing experiment. Ensure that no less than 90% of the overall stem cell culture remains pluripotent and have not spontaneously differentiated. Follow our complete step-by-step human iPS cell culture protocol guide including ECM coating, thawing, culturing and freezing of iPSCs prior to starting a CRISPR experiment.
Gene Editing of Human iPS Cells
This protocol is based on using 1 x 106 cells per nucleofection. This protocol is for two samples. For more samples, adjust the following protocol as necessary. Where targeted integration is desired, donor molecules must be designed and included in the transfections as well, and will require specific detection assays to determine integration efficiency. These methods will be governed by overall cleavage efficiency of the nuclease, distance from the cut site to the desired mutation site, and local sequence composition. For best results, it is highly recommended that the end user optimizes donor designs based on these criteria.
Basic Gene Editing Protocol
- Enzymatically detach human iPS cells from culture surface using Accutase (A6964) with Rock inhibitor, 10 µM (SCM075). When cells are ~90% confluent, each well of a 6-well plate will contain approximately 1.5 to 2 x 106 cells.
- Transfer cell suspension to a 15 mL conical tube and centrifuge at 200xg for 5 min.
- Open Lonza P3 kit (V4XP-3024). Add 200 µL of P3 solution to a clean 1.5 mL Eppendorf tube. Then add 44 µL of P3 supplement to the 200 µL of P3 solution.
- Gently mix. Aliquot 50 µL to an Eppendorf tube. Repeat for a second Eppendorf tube.
- For tube #1, add 2 µL pmaxGFP plasmid (2 µg total, Lonza kit) as nucleofection control.
- For tube #2 (and subsequent tubes), add 2-10 µg of test nucleic acid (CAS9 RNA, gRNA or DNA plasmids).
- Following centrifugation, aspirate media from cell pellet.
- Add 100 µL of P3 supplemented solution from step 3 to cell pellet and gently resuspend pellet.
- Aliquot 50 µL of cell suspension to tube #1 and mix by pipetting up and down 2-3 times. Transfer all contents to a labelled Lonza 4D cuvette.
- Repeat with tube #2.
- Perform nucleofection with either of these two conditions.
a. P3 solution: CM100 (20-50% survival, >90% nucleofection efficiency)
b. P3 solution: CM130 (50-75% survival, >75% nucleofection efficiency) - After adding nucleofection solution, allow cuvettes to incubate for 10 min. This step increases the nucleofection efficiency, but must be limited to 10 minutes increases the risk of cell death.
- At 10 min, transfer cell suspension to iPSC Growth Media+ Rock inhibitor, 10 uM (SCM075)
a. For maximal recovery when nucleofecting with CM100, it is advisable to plate all 1 x 10^6 cells onto a single well of an ECM coated plate.
b. For CM130 conditions, cells may be plated into 1 or 2 wells of a 6-well plate. - Incubate overnight in a 37 °C 5% CO2 humidified incubator.
- Exchange media with iPSC Growth Media and continue with normal iPS cell culture protocols.
Complex Gene Editing Protocol
For difficult to edit cell lines, we recommend the use of a 3-part synthetic CRISPR system with purified recombinant Cas9 protein and synthetic gRNA which is divided into a tracrRNA and a crRNA. The crRNA is variable and complementary to the target of interest, while the tracrRNA sequence is static.
- Resuspend 250 µg of recombinant CAS9 protein (CAS9PROT) in 50 uL nuclease-free water (W4502) for a 5µg/µL stock solution. Add 2 µL to reaction for 10 µg of CAS9 protein and store on ice.
- Target Complex Ratio is 4:4:1 (tracrRNA:crRNA:Cas9 protein)
- Dilute 2 nmol crRNAs diluted in 66uL nuclease-free water (W4502), 5 mmol tracrRNA diluted in 166ul nuclease-free water (W4502), and combine 8 uL of each crRNA and tracrRNA to achieve a 240 pmole concentration.
- Heat the 16 µL of crRNA/tracrRNA combo to 95 °C for 5 minutes, then cool to 4 °C or just place on ice.
- Add 10µg of Cas9 protein to your cooled crRNA/tracrRNA combo, mix, then incubate on ice for 20 minutes to allow the CRISPR complexes to form.
- CRISPR is ready to nucleofect using the protocol above. Nuclease delivery method is identical for ZFN and CRISPR reagents.
Assessing the Cleavage Efficiency of the Targeted Nuclease
Following transfection of the nuclease reagents, cells should be incubated for 24-72 hours before assessment of nuclease activity. The SURVEYOR nuclease assay is a widely accepted method to determine nuclease efficiency. Protocols for this method do not vary between ZFN and CRISPR nuclease formats, and a detailed method for this assay can be found in pages 5-8 of the CompoZr Custom Zinc Finger Nuclease Technical Bulletin.
Clone Isolation and Screening
It is highly recommended that clonal isolates be generated for genotypic screening in order to obtain pure populations of either knock-out cells or SNP converted cells. Limiting dilution and FACS are among the most common methods for cloning, and we recommend optimization of the cloning conditions for your desired cell line prior to attempting gene editing.
Tips and Tricks
- iPSC culture conditions: In order to successfully CRISPR edit human iPSCs, the cells must be highly undifferentiated and in the log phase of growth. It is recommended that the cells are >90% positive for pluripotency markers (Oct-4/Nanog) before initiating a gene editing experiment. The addition of ROCKi during passaging and gene editing greatly help in overall iPS cell survival.
- Optimized CRISPR design: CRISPR endonucleases have shown wide variation in their activity, even among multiple CRISPRs designed within close genomic proximity. For this reason, we highly recommend that you test 3 to 4 CRISPR nucleases that target different DNA sequences. Many online tools are available for CRISPR design, however, we have applied our core capabilities in specific ZFN design to the CRISPR/Cas system to create an in silico collection of genome-wide CRISPR target sequences.
- Efficient CRISPR delivery: Common transfection reagents can be used to introduce CRISPR DNA/RNA plasmids into iPSCs. However, we have shown that nucleofection is a robust delivery method for a wide variety of cell types including human iPSCs. For initial experiments in human cells, we advise nucleofecting maxi-prepped CRISPR plasmid DNA into well-validated cell types such as K562 or U2OS to assess double strand break activity or donor integration levels. Additionally, our single vector format contains a GFP-linked expression cassette for Cas9, so that transfected cells can be subsequently inspected by microscopy or FACS analysis to monitor transfection efficiency
- Increasing single cell cloning survival: In order to enhance single cell cloning success, we recommended adding ROCK inhibitor to the culture media during passaging and editing steps. Using MEF conditioned media can also enhance cell survival of human iPSCs at single cell levels. Transfection and nucleofection conditions also need to be optimized to prevent iPSC toxicity. Since our CRISPR plasmids contains a GFP-linked Cas9 expression cassette, fluorescence-activated cell sorting (FACS) can be used to isolate cell populations. We suggest that cells be sorted into fractions with low, medium, and high GFP expression levels prior to single cell isolation.
- Reagent handling and storage: Store CRISPR plasmid at –20 °C immediately upon arrival. Store CRISPR RNA at –80 °C immediately upon arrival. Please avoid repeated freeze thawing of the plasmid or RNA. The reagents can be stored at the recommended temperatures for up to 12 months. Practice aseptic technique to avoid DNase contamination of the components. Keep reagent vials and sample tubes closed when not in use.
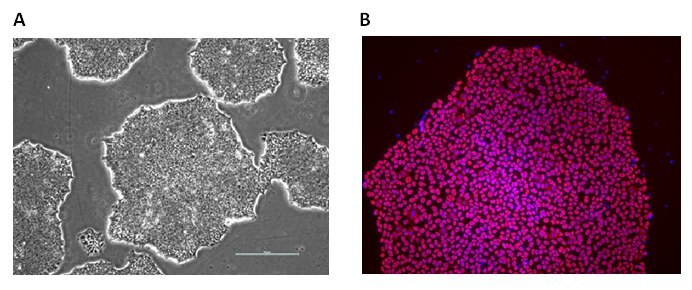
Figure 2. Alzheimer’s Human iPS Cell Lines. ApoE polymorphic alleles are the principal genetic determinants of Alzheimer disease (AD) risk. The EBiSC stem cell bank contains a complete set of isogenic lines, CRISPR engineered by Bioneer A/S, with the main ApoE genotypes: ApoE 2/2 (BIONi010-C-6), ApoE 3/3 (BIONi010-C-2) and ApoE 4/4 (BIONi010-C-1) as well as an ApoE knockout line (BIONi010-C-3) and TREM2 gene knockouts with homozygous R47H SNPs (BIONi010-C-7) or a homozygous T66M SNPs (BIONi010-C-8). A) iPS cells display normal undifferentiated phenotypes with colonies having clear defined borders and B) express the pluripotency marker Oct-4 (B).
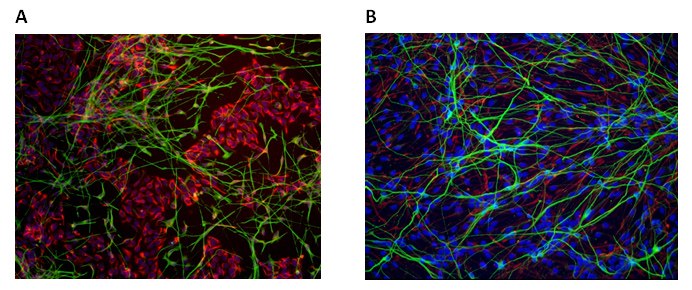
Figure 3. Neural Differentiation of Human iPS Cell Lines. CRISPR engineered human iPS cells can be differentiated into neural phenotypes with Nestin (A, red) and b-Tubulin (A, green) expression or glial phenotypes with GFAP (B, green) expression.
CRISPR Edited iPS Cell Lines |
|---|
To continue reading please sign in or create an account.
Don't Have An Account?