Use of PEPscreen® Peptides to Investigate Protein Farnesyltransferase Substrate Specificity
Prenylation is an essential post-translational modification for the proper localization and function of many proteins1,2, and is catalyzed by protein farnesyltransferase (FTase) and protein geranylgeranyltransferase type I (GGTase-I). These enzymes catalyze the covalent attachment of a 15-carbon farnesyl group from farnesyl diphosphate (FPP) or a 20-carbon geranylgeranyl group from geranylgeranyl diphosphate (GGPP) to the thiol side chain of a cysteine residue near the C-terminus of a protein substrate 2,3. The attached lipid aids in localization of proteins to cellular membranes and enhances protein-protein interactions4,5, and is required for the proper function of many proteins including members of the Ras and Rho superfamilies of small GTPases 1,6. While many proteins have been experimentally shown to be prenylated in vivo 7-10, the extent of prenylation within the proteome remains unclear. Based on comparison of known prenylated proteins, both FTase and GGTase-I have been proposed to recognize protein or peptide substrates containing a C-terminal “Ca1a2X” sequence 11-17. In this model, “C” refers to a cysteine three residues removed from the C-terminus that is prenylated at the thiol group to form a thioether, “a” refers to any aliphatic amino acid, and “X” refers to a subset of amino acids that are proposed to determine specificity for FTase (methionine, serine, glutamine, alanine) or GGTase-I (leucine, phenylalanine). Expanding upon the “Ca1a2X” box paradigm, bioinformatic analysis and biochemical studies of known substrates and related proteins indicate that sequences immediately upstream of the conserved cysteine residue may also play a role in substrate selectivity 8,18,19.
Prenylation has received significant interest as a target for pharmaceutical development, with FTase inhibitors (FTIs) in development as therapeutics for cancer, parasitic infection, and several other medical conditions 20-22. However, as the total complement of prenylated proteins is unknown, the FTase substrates responsible for FTI efficacy are not yet understood. Defining the prenylation-dependent pathways potentially responsible for the efficacy of these treatments could provide valuable insights for development of novel pharmaceuticals. Understanding the in vivo substrate selectivity of FTase and GGTase-I constitutes an important step towards characterizing the prenylated proteins involved in these pathways. Towards this goal, we have used peptide libraries to study the potential reactivity of human proteins with FTase, as well as to investigate the specific amino acid properties recognized by FTase within the Ca1a2X motif.
To identify potential novel prenylated proteins, we used a PEPscreen peptide library [Sigma Custom Products] to probe the reactivity of naturally occurring Ca1a2X sequences with FTase 23. To generate this library, we searched the human genome database for proteins that contain a cysteine four amino acids from the C-terminus as a minimal specificity requirement. From the resulting list of ~600 proteins, we selected approximately half to screen for activity with FTase. Peptides corresponding to the last four amino acids of these candidate substrates along with N-terminal Thr and Lys residues (TKCxxx) were screened for farnesylation by FTase under both kcat / Km peptide (multiple-turnover-, [Enzyme] << [Substrate]) and single-turnover ([Enzyme] > [Substrate]) reaction conditions. Out of the >300 peptides screened, FTase catalyzed multipleturnover farnesylation of 106 peptides, consistent with the parent proteins serving as FTase substrates in vivo. Surprisingly, of the remaining peptides, 67 % were farnesylated under single-turnover conditions, suggesting that there are two classes of substrates for FTase with distinct reactivity profiles. Analysis of the sequence preferences for these two classes of substrates illustrates a significant difference in FTase selectivity for the multiple- and single-turnover substrates, with the multiple-turnover substrates adhering closely to the current Ca1a2X model for FTase selectivity while the singleturnover substrates show distinct sequence preferences. These different classes of FTase substrates may reflect an unanticipated mechanism for regulating farnesylation, with potential impact on the localization, trafficking, and activity of prenylated proteins within the cell. These results improve the definition of prenyltransferase substrate specificity, test the efficacy of substrate algorithms, and provide valuable information about therapeutic targets. Finally, these data illuminate the potential for in vivo regulation of prenylation through modulation of single- versus multiple-turnover peptide reactivity with FTase.
Biochemical studies of prenyltransferase substrate specificity indicate that recognition of peptide substrates is more complex than originally proposed. For instance, although FTase and GGTase-I specificity is determined predominantly by the X residue 11,13-17,24-27, some substrates react efficiently with both enzymes 11,27. Peptide substrate specificity also depends on interactions of the peptide with the FPP co-substrate 28-30. As these findings reflect, understanding FTase and GGTase-I substrate specificity will require identification and energetic characterization of interactions involved in substrate recognition. In a second study, we used PEPscreen peptides and applied structure-function analysis to define the specific selectivity criteria that FTase employs to recognize the a2 residue of substrate peptides31. We measured the reactivity of FTase with several panels of peptides in which the a2 residue is substituted with all twenty amino acids, while keeping the remainder of the Ca1a2X sequence constant. Correlation of peptide reactivity within each panel against both the amino acid polarity and steric volume of the a2 residue indicates that FTase recognizes both the size and hydrophobicity of the residue at the a2 position, contrasting with the predominantly polarity-based recognition observed at the X residue 19. Furthermore, comparison across peptide panels indicates that a2 selectivity is also affected by the identity of the adjacent X residue, leading to context-dependent substrate recognition. These findings suggest that the current model describing FTase selectivity reflects only a subset of potential FTase substrates, raising the possibility of novel FTase substrates whose C-terminal sequences do not conform to the canonical Ca1a2X motif.
Muliple turn-over sequences |
|---|
Peptides are a form of dansyl-TKCxxx, where x represents any amino acid.
Peptides exhibited multiple turn-over reactivity in initial screens using either fluorescence or radioactive detection.
Single turn-over sequences |
|---|
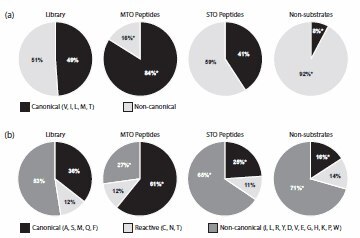
Figure 1.Amino acid compositions at the a2 and X positions of the Ca1a2X sequence in the initial library. (A) Amino acid compositions at the a2 position of the Ca1a2X sequence. The percentages of amino acids at the a2 position, grouped into either canonical (V, I, L, M, and T) or non-canonical residues, are plotted for the initial library, the MTO substrates, the STO substrates, and the non-substrate peptides. Wedges are labeled with the percentage representation of the classes of amino acids within each pool of peptides. An asterisk (*) denotes those percentages that are significantly different in the substrate pools as compared to the library (p<0.02). (B) Amino acid compositions at the X position of the Ca1a2X sequence. The percentages of amino acids at the X position, grouped into canonical (A, S, M, Q, and F), reactive (C, N, and T), and non-canonical residues, are plotted for the initial library, the MTO substrates, the STO substrates, and the non-substrate peptides. Wedges are labeled with the percentage representation of the classes of amino acids within each pool of peptides; percentages may not add to 100% due to rounding. An asterisk (*) denotes those percentages that are significantly different in the substrate pools as compared to the library (p<0.02).
References
To continue reading please sign in or create an account.
Don't Have An Account?