A Quantitative Mass Spectrometry-based Approach for Identifying Protein Kinase Clients and Quantifying Kinase Activity
Y. Huang, N. L. Houston, A. Tovar-Mendez, S. E. Stevenson, J. A. Miernyk, D. D. Randall, J. J. Thelen
Huang et al.1 have recently developed a quantitative mass spectrometry-based assay for identifying protein kinase substrates and quantifying kinase activity. This approach offers several advantages over current methods:
- It is label-free and eliminates the need for fluorescent or radiolabel detection of phosphorylated substrates
- It is solution-based, which prevents the protein from adopting non-native conformations and non-specific binding of the probe to the chip
- It uses tandem mass spectrometry (MS/MS) to both identify kinase-substrate proteins and to quantify kinase activity in a quick, sensitive, and reproducible manner
Pyruvate dehydrogenase kinase (PDK) was chosen as the model for in vitro assays. The peptide substrate and the phosphorylated peptide product were separated and identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Comparison of multistage activation (MSA)2,3 and other MS/MS ion dissociation strategies indicated that, for some peptides, MSA was a more robust method for phosphopeptide analysis, allowing the peptide to be identified with a higher degree of confidence. The specificity of the assay was demonstrated by incubating a synthetic peptide cocktail, containing 46 peptides (PEPscreen®, Custom Products), covering all Ser, Thr, and Tyr residues in the substrate protein, pyruvate dehydrogenase alpha subunit, and the kinase itself with recombinant Arabidopsis thaliana PDK (AtPDK); (YHGHSMSDPGSTYR) was the only substrate to be phosphorylated (Figure 1). In the absence of kinase or when Met293 (which when converted to MetSO acts as an inhibitor of Ser292 phosphorylation) was chemically oxidized, no phosphorylation was observed (Figure 2), illustrating the sensitivity of the assay. Substitution analysis of the PDK substrate was performed using a series of peptide variants synthesized on PEPscreen, and it revealed the importance of key amino acid residues in retaining the kinase activity (Figure 3). The assay was also tested using a calciumdependent protein kinase (CPK3) against a known substrate (Figure 2). In the absence of calcium chloride, on which CPK3 is dependent, phosphorylation was abolished (Figure 2).
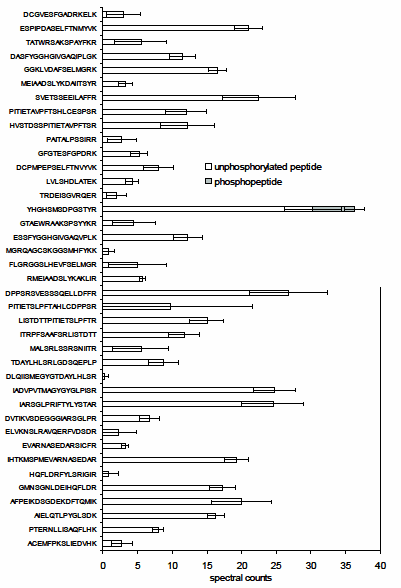
Figure 1.The in vitro assay based upon spectral counting4,5 reveals kinase specificity in peptide cocktails (PEPscreen, Custom Products). The recombinant AtPDK phosphorylates only YHGHSMSDPGSTYR in the peptide cocktail, which contains all potential Ser-containing tryptic peptides derived from AtPDC E1α and AtPDK. All values are means ± std of four repeats.
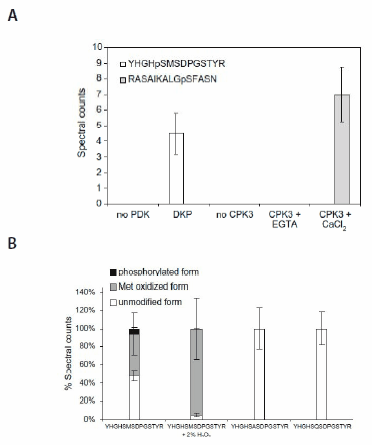
Figure 2. The in vitro assay reveals kinase sensitivity. (A) The recombinant PDK and CPK3 phosphorylate respective cognate substrate peptide in cocktail. (B) Oxidation of Met293 inhibits phosphorylation of Ser292 in the peptide substrate YHGHSMSDPGSTYR. All values are means ± std of four repeats.
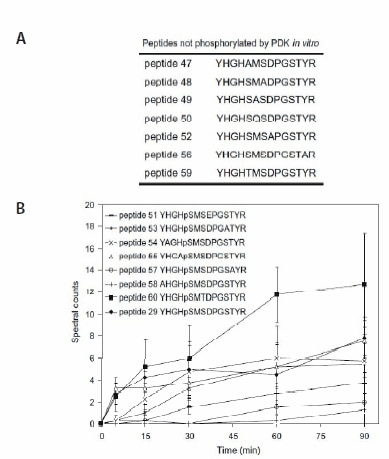
Figure 3. In vitro assays containing peptide variants of YHGHSMSDPGSTYR (PEPscreen, Custom Products). (A) Peptide variants not phosphorylated by PDK. (B) Spectral counts of peptides phosphorylated by AtPDK as a function of reaction time. All values are means ± std of four repeats.
Spectral counting4,5 based on LC-MS/MS allowed the authors to monitor the in vitro activity of PDK by quantifying the relative abundance of both the substrate and the phosphorylated product. The peptides were separated by liquid chromatography (LC) before identification and quantified using the MS/MS–MSA method. The kinetic constants of the reaction were calculated based on these data (Figure 4). The data followed nonMichaelis– Menten kinetics, showing reduced PDK activity, at substrate peptide concentrations above 180 μmol/L. This suggests that the reaction product (phosphorylated peptide) acts as a PDK inhibitor. In conclusion, the use of LC-MS/MS combined with convenient access to affordable peptide libraries yields a promising approach to the identification and quantification of protein kinase enzymatic activity.
![A quantitative mass spectrometry-based approach for identifying protein kinase clients and quantifying kinase activity Initial rate analysis of AtPDK. (A) Progress curve of PDK activity. (B) Dynamic range of detection and quantitation of substrate and product peptides. (C) Plot of V versus [S]. All values are means ± std of four repeats. rPDK, recombinant pyruvate dehydrogenase kinase.](/deepweb/assets/sigmaaldrich/marketing/global/images/technical-documents/articles/analytical-chemistry/large-molecule-hplc/rateanalysis.gif)
Figure 4. Initial rate analysis of AtPDK. (A) Progress curve of PDK activity. (B) Dynamic range of detection and quantitation of substrate and product peptides. (C) Plot of V versus [S]. All values are means ± std of four repeats. rPDK, recombinant pyruvate dehydrogenase kinase.
References
To continue reading please sign in or create an account.
Don't Have An Account?